KPCs
Klebsiella pneumoniaecarbapenemases (KPCs) result in resistance to virtually all β-lactam antibiotics, leaving very few therapeutic options.
KPCs are plasmid-encoded Amber class A carbapenemases that were first identified in K pneumoniae isolates in North America in 1996. At present, plasmids carrying the gene encoding for KPC have been found in other Enterobacteriaceae species including Escherichia coli.
NDM-1 Carbapenemases
Organisms carrying the NDM-1 β-lactamase are resistant to essentially all β-lactam drugs.
New Delhimetallo-beta-lactamase-1 (NDM-1) is a type of carbapenemase that was first reported in 2009 when found in a K pneumoniae strain and an E coli strain from a Swedish patient repatriated from a New Delhi hospital. Many other cases have been reported since, indicating rapid spread of these strains. Fortunately, NDM-1 isolates are not common in the U.S.
Extended Spectrum Beta-Lactamases (ESBLs)
TEM, SHV, and CTX-M b-lactamases are generally capable of hydrolyzing penicillins, monobactams, and cephalosporins of the first, second, third, and fourth generations, thereby limiting treatment options.
The TEM, SHV, and CTX-M b-lactamases are the clinically most significant plasmid-encoded extended spectrum beta-lactamases (ESBL). TEM and SHV are the result of point mutations leading to amino acid substitutions. These amino acid substitutions cause hydrolysis of a wide range of β-lactam antibiotics. CTX-M b-lactamases originate in Kluyvera species and presently are the most prevalent ESBLs. Based on amino acid sequence, they can be divided into at least 5 different families: CTX-M1, CTX-M2, CTX-M9, CTX-M8 and CTX-M25.
AmpC Cephalosporinases
The AmpC cephalosporinases are able to hydrolyze almost all β-lactam antibiotics including the penicillins, cephalosporins, and monobactams. Unlike ESBLs, AmpC cephalosporinases are not inhibited by clavulanic acid. Organisms harboring AmpC cephalosporinases normally remain susceptible to the carbapenems (imipenem, meropenem, doripenem) and 4th generation cephalosporins such as cefepime.
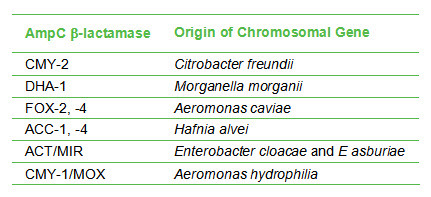
Generally, AmpC-encoding genes carried on plasmids give rise to significantly increased β-lactamase production compared to the chromosomally-located genes. This implies that organisms with plasmid-encoded β-lactamase are more resistant to antibiotics; hence it is useful to discriminate between organisms with plasmid-encoded vs chromosomal-encoded AmpC β-lactamases. Species that carry chromosomal-encoded AmpC β-lactamases are shown in the table below. Plasmid-encoded AmpC β-lactamases are found in species other than those listed in the table, ie, those that don’t carry chromosomal-encoded AmpC b-lactamases. E coli is an example of a species that carries plasmid-encoded AmpC β-lactamases. Additionally, the presence of a plasmid-encoded enzyme is indicated when an AmpC β-lactamase type is found in an organism not normally associated with that type. For example, Morganella morganii is normally associated with DHA-1. Detection of DHA-1 in a species other than M morganii could indicate the presence of a plasmid-encoded AmpC β-lactamase.
Your Privacy Choices





